Info
cellxgene_
Wilson et al. (2022)
1.28 GiB
02-02-2024
39176 cells × 27980 genes
Quick links
Used in
No related benchmarks found.
Multimodal single cell sequencing implicates chromatin accessibility and genetic background in diabetic kidney disease progression
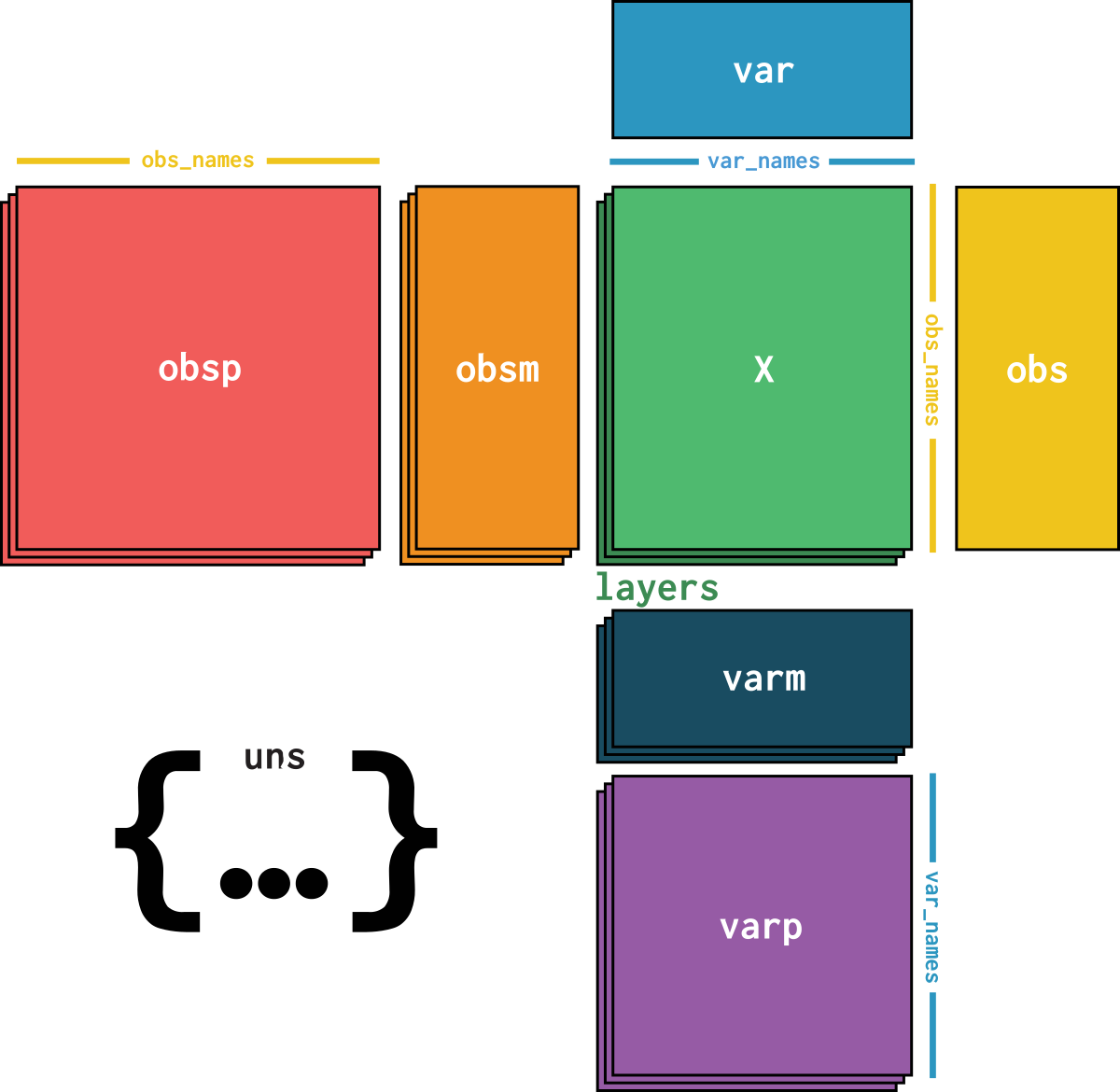
CREATED
02-02-2024
DIMENSIONS
39176 × 27980
Multimodal single cell sequencing is a powerful tool for interrogating cell-specific changes in transcription and chromatin accessibility. We performed single nucleus RNA (snRNA-seq) and assay for transposase accessible chromatin sequencing (snATAC-seq) on human kidney cortex from donors with and without diabetic kidney disease (DKD) to identify altered signaling pathways and transcription factors associated with DKD. Both snRNA-seq and snATAC-seq had an increased proportion of VCAM1+ injured proximal tubule cells (PT_VCAM1) in DKD samples. PT_VCAM1 has a pro-inflammatory expression signature and transcription factor motif enrichment implicated NFkB signaling. We used stratified linkage disequilibrium score regression to partition heritability of kidney-function-related traits using publicly-available GWAS summary statistics. Cell-specific PT_VCAM1 peaks were enriched for heritability of chronic kidney disease (CKD), suggesting that genetic background may regulate chromatin accessibility and DKD progression. snATAC-seq found cell-specific differentially accessible regions (DAR) throughout the nephron that change accessibility in DKD and these regions were enriched for glucocorticoid receptor (GR) motifs. Changes in chromatin accessibility were associated with decreased expression of insulin receptor, increased gluconeogenesis, and decreased expression of the GR cytosolic chaperone, FKBP5, in the diabetic proximal tubule. Cleavage under targets and release using nuclease (CUT&RUN) profiling of GR binding in bulk kidney cortex and an in vitro model of the proximal tubule (RPTEC) showed that DAR co-localize with GR binding sites. CRISPRi silencing of GR response elements (GRE) in the FKBP5 gene body reduced FKBP5 expression in RPTEC, suggesting that reduced FKBP5 chromatin accessibility in DKD may alter cellular response to GR. We developed an open-source tool for single cell allele specific analysis (SALSA) to model the effect of genetic background on gene expression. Heterozygous germline single nucleotide variants (SNV) in proximal tubule ATAC peaks were associated with allele-specific chromatin accessibility and differential expression of target genes within cis-coaccessibility networks. Partitioned heritability of proximal tubule ATAC peaks with a predicted allele-specific effect was enriched for eGFR, suggesting that genetic background may modify DKD progression in a cell-specific manner.
dataset is an AnnData object with n_obs × n_vars = 39176 × 27980 with slots:
soma_joinid, dataset_id, assay, assay_ontology_term_id, cell_type, cell_type_ontology_term_id, development_stage, development_stage_ontology_term_id, disease, disease_ontology_term_id, donor_id, is_primary_data, self_reported_ethnicity, self_reported_ethnicity_ontology_term_id, sex, sex_ontology_term_id, suspension_type, tissue, tissue_ontology_term_id, tissue_general, tissue_general_ontology_term_id, batch, size_factorssoma_joinid, feature_id, feature_name, hvg, hvg_scoreknn_connectivities, knn_distancesX_pcapca_loadingscounts, normalizeddataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url, knn, normalization_id, pca_variance| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| obs | ||||
assay
|
Type of assay used to generate the cell data, indicating the methodology or technique employed. |
vector
|
category
|
39176 |
assay_
|
Experimental Factor Ontology (EFO:) term identifier for the assay, providing a standardized reference to the assay type.
|
vector
|
category
|
39176 |
batch
|
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc. |
vector
|
category
|
39176 |
cell_
|
Classification of the cell type based on its characteristics and function within the tissue or organism. |
vector
|
category
|
39176 |
cell_
|
Cell Ontology (CL:) term identifier for the cell type, offering a standardized reference to the specific cell classification.
|
vector
|
category
|
39176 |
dataset_
|
Identifier for the dataset from which the cell data is derived, useful for tracking and referencing purposes. |
vector
|
category
|
39176 |
development_
|
Stage of development of the organism or tissue from which the cell is derived, indicating its maturity or developmental phase. |
vector
|
category
|
39176 |
development_
|
Ontology term identifier for the developmental stage, providing a standardized reference to the organism’s developmental phase. If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Developmental Stages (HsapDv:) ontology is used. If the organism is mouse (organism_ontology_term_id == 'NCBITaxon:10090'), then the Mouse Developmental Stages (MmusDv:) ontology is used. Otherwise, the Uberon (UBERON:) ontology is used.
|
vector
|
category
|
39176 |
disease
|
Information on any disease or pathological condition associated with the cell or donor. |
vector
|
category
|
39176 |
disease_
|
Ontology term identifier for the disease, enabling standardized disease classification and referencing. Must be a term from the Mondo Disease Ontology (MONDO:) ontology term, or PATO:0000461 from the Phenotype And Trait Ontology (PATO:).
|
vector
|
category
|
39176 |
donor_
|
Identifier for the donor from whom the cell sample is obtained. |
vector
|
category
|
39176 |
is_
|
Indicates whether the data is primary (directly obtained from experiments) or has been computationally derived from other primary data. |
vector
|
bool
|
39176 |
self_
|
Ethnicity of the donor as self-reported, relevant for studies considering genetic diversity and population-specific traits. |
vector
|
category
|
39176 |
self_
|
Ontology term identifier for the self-reported ethnicity, providing a standardized reference for ethnic classifications. If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Ancestry Ontology (HANCESTRO:) is used.
|
vector
|
category
|
39176 |
sex
|
Biological sex of the donor or source organism, crucial for studies involving sex-specific traits or conditions. |
vector
|
category
|
39176 |
sex_
|
Ontology term identifier for the biological sex, ensuring standardized classification of sex. Only PATO:0000383, PATO:0000384 and PATO:0001340 are allowed.
|
vector
|
category
|
39176 |
size_
|
The size factors created by the normalisation method, if any. |
vector
|
float32
|
39176 |
soma_
|
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the cell. |
vector
|
int64
|
39176 |
suspension_
|
Type of suspension or medium in which the cells were stored or processed, important for understanding cell handling and conditions. |
vector
|
category
|
39176 |
tissue
|
Specific tissue from which the cells were derived, key for context and specificity in cell studies. |
vector
|
category
|
39176 |
tissue_
|
General category or classification of the tissue, useful for broader grouping and comparison of cell data. |
vector
|
category
|
39176 |
tissue_
|
Ontology term identifier for the general tissue category, aiding in standardizing and grouping tissue types. For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
|
vector
|
category
|
39176 |
tissue_
|
Ontology term identifier for the tissue, providing a standardized reference for the tissue type. For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
|
vector
|
category
|
39176 |
| var | ||||
feature_
|
Unique identifier for the feature, usually a ENSEMBL gene id. |
vector
|
object
|
27980 |
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
27980 |
hvg
|
Whether or not the feature is considered to be a ‘highly variable gene’ |
vector
|
bool
|
27980 |
hvg_
|
A ranking of the features by hvg. |
vector
|
float64
|
27980 |
soma_
|
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the feature. |
vector
|
int64
|
27980 |
| obsp | ||||
knn_
|
K nearest neighbors connectivities matrix. |
sparsematrix
|
float32
|
39176 × 39176 |
knn_
|
K nearest neighbors distance matrix. |
sparsematrix
|
float64
|
39176 × 39176 |
| obsm | ||||
X_
|
The resulting PCA embedding. |
densematrix
|
float32
|
39176 × 50 |
| varm | ||||
pca_
|
The PCA loadings matrix. |
densematrix
|
float32
|
27980 × 50 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
39176 × 27980 |
normalized
|
Normalised expression values |
sparsematrix
|
float32
|
39176 × 27980 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
knn
|
Supplementary K nearest neighbors data. |
dict
|
3 | |
normalization_
|
Which normalization was used |
atomic
|
str
|
1 |
pca_
|
The PCA variance objects. |
dict
|
2 | |
dataset.layers['counts']In R: dataset$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 39176 × 27980
Raw counts
dataset.layers['normalized']In R: dataset$layers[["normalized"]]
Type: sparsematrix, data type: float32, shape: 39176 × 27980
Normalised expression values
dataset.obs['soma_joinid']In R: dataset$obs[["soma_joinid"]]
Type: vector, data type: int64, shape: 39176
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the cell.
dataset.obs['dataset_id']In R: dataset$obs[["dataset_id"]]
Type: vector, data type: category, shape: 39176
Identifier for the dataset from which the cell data is derived, useful for tracking and referencing purposes.
dataset.obs['assay']In R: dataset$obs[["assay"]]
Type: vector, data type: category, shape: 39176
Type of assay used to generate the cell data, indicating the methodology or technique employed.
dataset.obs['assay_ontology_term_id']In R: dataset$obs[["assay_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Experimental Factor Ontology (EFO:) term identifier for the assay, providing a standardized reference to the assay type.
dataset.obs['cell_type']In R: dataset$obs[["cell_type"]]
Type: vector, data type: category, shape: 39176
Classification of the cell type based on its characteristics and function within the tissue or organism.
dataset.obs['cell_type_ontology_term_id']In R: dataset$obs[["cell_type_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Cell Ontology (CL:) term identifier for the cell type, offering a standardized reference to the specific cell classification.
dataset.obs['development_stage']In R: dataset$obs[["development_stage"]]
Type: vector, data type: category, shape: 39176
Stage of development of the organism or tissue from which the cell is derived, indicating its maturity or developmental phase.
dataset.obs['development_stage_ontology_term_id']In R: dataset$obs[["development_stage_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Ontology term identifier for the developmental stage, providing a standardized reference to the organism’s developmental phase.
If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Developmental Stages (HsapDv:) ontology is used.
If the organism is mouse (organism_ontology_term_id == 'NCBITaxon:10090'), then the Mouse Developmental Stages (MmusDv:) ontology is used. Otherwise, the Uberon (UBERON:) ontology is used.
dataset.obs['disease']In R: dataset$obs[["disease"]]
Type: vector, data type: category, shape: 39176
Information on any disease or pathological condition associated with the cell or donor.
dataset.obs['disease_ontology_term_id']In R: dataset$obs[["disease_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Ontology term identifier for the disease, enabling standardized disease classification and referencing.
Must be a term from the Mondo Disease Ontology (MONDO:) ontology term, or PATO:0000461 from the Phenotype And Trait Ontology (PATO:).
dataset.obs['donor_id']In R: dataset$obs[["donor_id"]]
Type: vector, data type: category, shape: 39176
Identifier for the donor from whom the cell sample is obtained.
dataset.obs['is_primary_data']In R: dataset$obs[["is_primary_data"]]
Type: vector, data type: bool, shape: 39176
Indicates whether the data is primary (directly obtained from experiments) or has been computationally derived from other primary data.
dataset.obs['self_reported_ethnicity']In R: dataset$obs[["self_reported_ethnicity"]]
Type: vector, data type: category, shape: 39176
Ethnicity of the donor as self-reported, relevant for studies considering genetic diversity and population-specific traits.
dataset.obs['self_reported_ethnicity_ontology_term_id']In R: dataset$obs[["self_reported_ethnicity_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Ontology term identifier for the self-reported ethnicity, providing a standardized reference for ethnic classifications.
If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Ancestry Ontology (HANCESTRO:) is used.
dataset.obs['sex']In R: dataset$obs[["sex"]]
Type: vector, data type: category, shape: 39176
Biological sex of the donor or source organism, crucial for studies involving sex-specific traits or conditions.
dataset.obs['sex_ontology_term_id']In R: dataset$obs[["sex_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Ontology term identifier for the biological sex, ensuring standardized classification of sex. Only PATO:0000383, PATO:0000384 and PATO:0001340 are allowed.
dataset.obs['suspension_type']In R: dataset$obs[["suspension_type"]]
Type: vector, data type: category, shape: 39176
Type of suspension or medium in which the cells were stored or processed, important for understanding cell handling and conditions.
dataset.obs['tissue']In R: dataset$obs[["tissue"]]
Type: vector, data type: category, shape: 39176
Specific tissue from which the cells were derived, key for context and specificity in cell studies.
dataset.obs['tissue_ontology_term_id']In R: dataset$obs[["tissue_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Ontology term identifier for the tissue, providing a standardized reference for the tissue type.
For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
dataset.obs['tissue_general']In R: dataset$obs[["tissue_general"]]
Type: vector, data type: category, shape: 39176
General category or classification of the tissue, useful for broader grouping and comparison of cell data.
dataset.obs['tissue_general_ontology_term_id']In R: dataset$obs[["tissue_general_ontology_term_id"]]
Type: vector, data type: category, shape: 39176
Ontology term identifier for the general tissue category, aiding in standardizing and grouping tissue types.
For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
dataset.obs['batch']In R: dataset$obs[["batch"]]
Type: vector, data type: category, shape: 39176
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc.
dataset.obs['size_factors']In R: dataset$obs[["size_factors"]]
Type: vector, data type: float32, shape: 39176
The size factors created by the normalisation method, if any.
dataset.obsm['X_pca']In R: dataset$obsm[["X_pca"]]
Type: densematrix, data type: float32, shape: 39176 × 50
The resulting PCA embedding.
dataset.obsp['knn_connectivities']In R: dataset$obsp[["knn_connectivities"]]
Type: sparsematrix, data type: float32, shape: 39176 × 39176
K nearest neighbors connectivities matrix.
dataset.obsp['knn_distances']In R: dataset$obsp[["knn_distances"]]
Type: sparsematrix, data type: float64, shape: 39176 × 39176
K nearest neighbors distance matrix.
dataset.uns['dataset_description']In R: dataset$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset.uns['dataset_id']In R: dataset$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset.uns['dataset_name']In R: dataset$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset.uns['dataset_organism']In R: dataset$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset.uns['dataset_reference']In R: dataset$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset.uns['dataset_summary']In R: dataset$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset.uns['dataset_url']In R: dataset$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset.uns['knn']In R: dataset$uns[["knn"]]
Type: dict, data type: ``, shape: 3
Supplementary K nearest neighbors data.
dataset.uns['normalization_id']In R: dataset$uns[["normalization_id"]]
Type: atomic, data type: str, shape: 1
Which normalization was used
dataset.uns['pca_variance']In R: dataset$uns[["pca_variance"]]
Type: dict, data type: ``, shape: 2
The PCA variance objects.
dataset.var['soma_joinid']In R: dataset$var[["soma_joinid"]]
Type: vector, data type: int64, shape: 27980
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the feature.
dataset.var['feature_id']In R: dataset$var[["feature_id"]]
Type: vector, data type: object, shape: 27980
Unique identifier for the feature, usually a ENSEMBL gene id.
dataset.var['feature_name']In R: dataset$var[["feature_name"]]
Type: vector, data type: object, shape: 27980
A human-readable name for the feature, usually a gene symbol.
dataset.var['hvg']In R: dataset$var[["hvg"]]
Type: vector, data type: bool, shape: 27980
Whether or not the feature is considered to be a ‘highly variable gene’
dataset.var['hvg_score']In R: dataset$var[["hvg_score"]]
Type: vector, data type: float64, shape: 27980
A ranking of the features by hvg.
dataset.varm['pca_loadings']In R: dataset$varm[["pca_loadings"]]
Type: densematrix, data type: float32, shape: 27980 × 50
The PCA loadings matrix.