Info
openproblems_
Luecken et al. (2021)
1.26 GiB
02-02-2024
16382 cells × 18771 genes
Human pancreas cells dataset from the scIB benchmarks
openproblems_
Luecken et al. (2021)
1.26 GiB
02-02-2024
16382 cells × 18771 genes
CREATED
02-02-2024
DIMENSIONS
16382 × 18771
Human pancreatic islet scRNA-seq data from 6 datasets across technologies (CEL-seq, CEL-seq2, Smart-seq2, inDrop, Fluidigm C1, and SMARTER-seq).
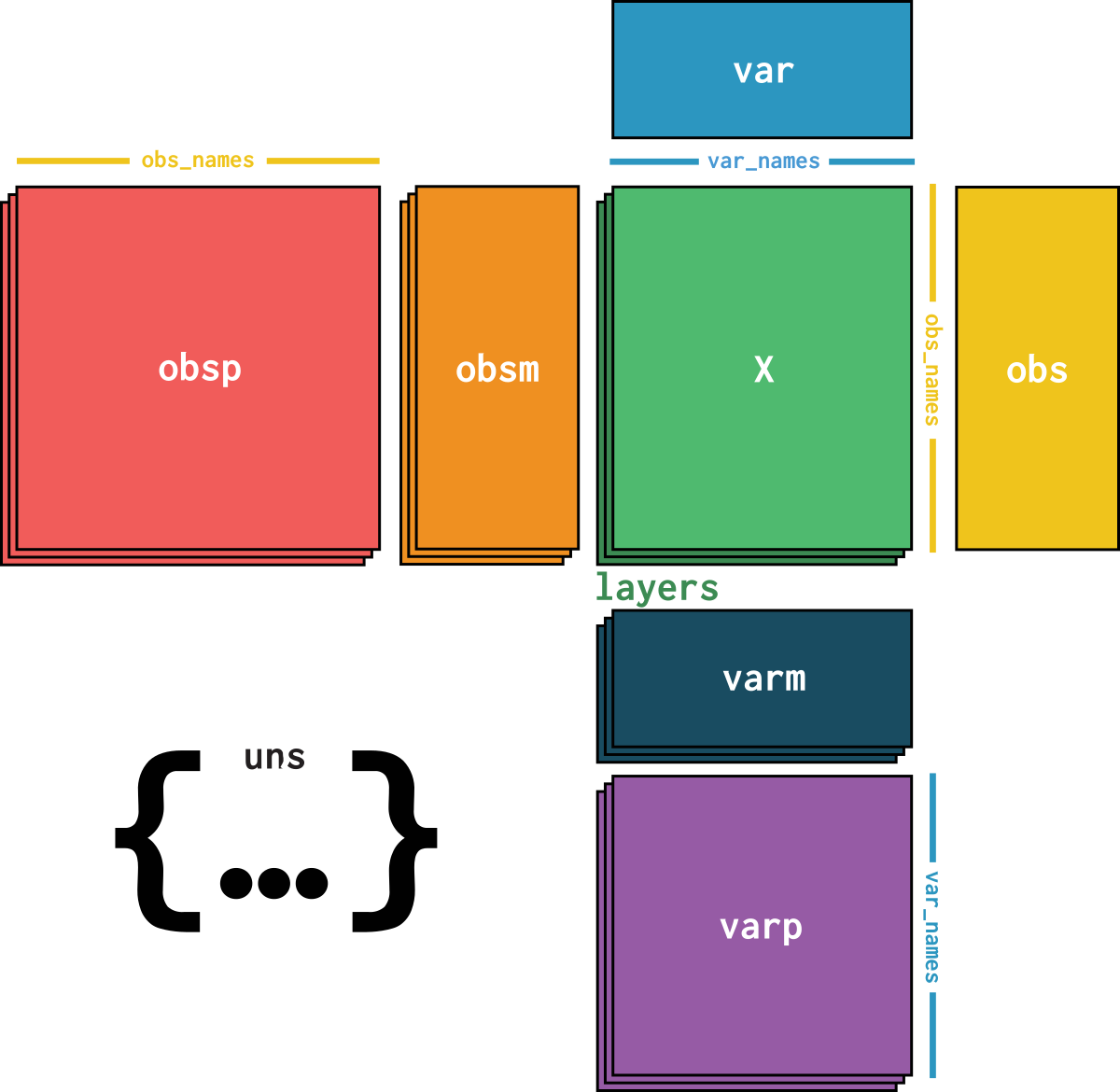
dataset is an AnnData object with n_obs × n_vars = 16382 × 18771 with slots:
size_factors, cell_type, batchfeature_name, hvg, hvg_scoreknn_connectivities, knn_distancesX_pcapca_loadingscounts, normalizeddataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url, knn, normalization_id, pca_variance| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| obs | ||||
batch
|
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc. |
vector
|
category
|
16382 |
cell_
|
Classification of the cell type based on its characteristics and function within the tissue or organism. |
vector
|
category
|
16382 |
size_
|
The size factors created by the normalisation method, if any. |
vector
|
float32
|
16382 |
| var | ||||
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
18771 |
hvg
|
Whether or not the feature is considered to be a ‘highly variable gene’ |
vector
|
bool
|
18771 |
hvg_
|
A ranking of the features by hvg. |
vector
|
float64
|
18771 |
| obsp | ||||
knn_
|
K nearest neighbors connectivities matrix. |
sparsematrix
|
float32
|
16382 × 16382 |
knn_
|
K nearest neighbors distance matrix. |
sparsematrix
|
float64
|
16382 × 16382 |
| obsm | ||||
X_
|
The resulting PCA embedding. |
densematrix
|
float32
|
16382 × 50 |
| varm | ||||
pca_
|
The PCA loadings matrix. |
densematrix
|
float64
|
18771 × 50 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
16382 × 18771 |
normalized
|
Normalised expression values |
sparsematrix
|
float32
|
16382 × 18771 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
knn
|
Supplementary K nearest neighbors data. |
dict
|
3 | |
normalization_
|
Which normalization was used |
atomic
|
str
|
1 |
pca_
|
The PCA variance objects. |
dict
|
2 | |
dataset.layers['counts']In R: dataset$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 16382 × 18771
Raw counts
dataset.layers['normalized']In R: dataset$layers[["normalized"]]
Type: sparsematrix, data type: float32, shape: 16382 × 18771
Normalised expression values
dataset.obs['size_factors']In R: dataset$obs[["size_factors"]]
Type: vector, data type: float32, shape: 16382
The size factors created by the normalisation method, if any.
dataset.obs['cell_type']In R: dataset$obs[["cell_type"]]
Type: vector, data type: category, shape: 16382
Classification of the cell type based on its characteristics and function within the tissue or organism.
dataset.obs['batch']In R: dataset$obs[["batch"]]
Type: vector, data type: category, shape: 16382
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc.
dataset.obsm['X_pca']In R: dataset$obsm[["X_pca"]]
Type: densematrix, data type: float32, shape: 16382 × 50
The resulting PCA embedding.
dataset.obsp['knn_connectivities']In R: dataset$obsp[["knn_connectivities"]]
Type: sparsematrix, data type: float32, shape: 16382 × 16382
K nearest neighbors connectivities matrix.
dataset.obsp['knn_distances']In R: dataset$obsp[["knn_distances"]]
Type: sparsematrix, data type: float64, shape: 16382 × 16382
K nearest neighbors distance matrix.
dataset.uns['dataset_description']In R: dataset$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset.uns['dataset_id']In R: dataset$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset.uns['dataset_name']In R: dataset$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset.uns['dataset_organism']In R: dataset$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset.uns['dataset_reference']In R: dataset$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset.uns['dataset_summary']In R: dataset$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset.uns['dataset_url']In R: dataset$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset.uns['knn']In R: dataset$uns[["knn"]]
Type: dict, data type: ``, shape: 3
Supplementary K nearest neighbors data.
dataset.uns['normalization_id']In R: dataset$uns[["normalization_id"]]
Type: atomic, data type: str, shape: 1
Which normalization was used
dataset.uns['pca_variance']In R: dataset$uns[["pca_variance"]]
Type: dict, data type: ``, shape: 2
The PCA variance objects.
dataset.var['feature_name']In R: dataset$var[["feature_name"]]
Type: vector, data type: object, shape: 18771
A human-readable name for the feature, usually a gene symbol.
dataset.var['hvg']In R: dataset$var[["hvg"]]
Type: vector, data type: bool, shape: 18771
Whether or not the feature is considered to be a ‘highly variable gene’
dataset.var['hvg_score']In R: dataset$var[["hvg_score"]]
Type: vector, data type: float64, shape: 18771
A ranking of the features by hvg.
dataset.varm['pca_loadings']In R: dataset$varm[["pca_loadings"]]
Type: densematrix, data type: float64, shape: 18771 × 50
The PCA loadings matrix.